Final Report/Thesis 2019: Difference between revisions
| (19 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
==Abstract== | ==Abstract== | ||
Somerton Man case is most mysterious case in | The Somerton Man's case is one of the most mysterious case in South Australia until today. An unknown man was murdered on the Somerton beach, and the identification of the victim is still a mystery. The project aims to investigate the identification of the Somerton man by means of studying his DNA data provided. Unfortunately, the DNA data was corrupted and has a high drop rate, therefore the project team was required to use different strategies and techniques to recover and analyse the DNA, then try to find out any possible characteristics of the Somerton Man. | ||
To approach the goals of the project, the team | To approach the goals of the project, the team firstly evaluates the DNA data and try to conduct DNA analysis via different genetic services. In addition, a degradation process of complete DNA data was done. By degrading several complete DNA files, we can observe how degradation could affect DNA analysis results. | ||
==Introduction== | ==Introduction== | ||
===Motivation=== | ===Motivation=== | ||
The main topic of the project is human identification | The main topic of the project is human identification using software programming and genetic analysis techniques. The project conducts a study on investigating the identification of the victim, Somerton Man, which is one of the most mysterious cases in South Australia until today. On December 1st 1948, a well-dressed male was found dead on Somerton Beach in Adelaide. He was clean-shaven, well dressed in a suit and no belongings of him that could prove his identity. Later the man was called as the Somerton Man. The figure below shows the look of the Somerton Man. | ||
[[File:SomertonMan.jpg|thumb|300px|center|Figure 1: The Somerton Man]] | [[File:SomertonMan.jpg|thumb|300px|center|Figure 1: The Somerton Man]] | ||
After more than half century, the identification of the Somerton Man is still | After more than half century, the identification of the Somerton Man is still unknown. With the Somerton Man's DNA data extracted from his hair, the project team may be able to conduct several DNA examinations and identify characteristics of the man. Unfortunately, the DNA data is corrupted due to degradation of the hair, but the team of the project will try the best to investigate the data with modern techniques. | ||
In modern society, human identification techniques is useful in multiple aspects, such as criminal investigation or seeking relatives. The most current identification techniques would require high quality DNA samples, but the project focus on investigating identification techniques based on low quality genetic data. Also the project concentrates on using engineering methods and problem solving skills to improve the identification techniques. | |||
===Objectives=== | ===Objectives=== | ||
The aim of the project is to investigate the identification of the Somerton Man. To be more specific, the group is aiming to identify any possible relatives, physical characteristics, genetic diseases or ethnicity of the Somerton Man. To achieve these goals, the team | The aim of the project is to investigate the identification of the Somerton Man. To be more specific, the group is aiming to identify any possible relatives, physical characteristics, genetic diseases or ethnicity of the Somerton Man. To achieve these goals, the team used software and genetic analysis techniques to work on the Somerton Man's DNA data. | ||
In addition, the reliability of DNA analysis results from a low quality DNA data would be investigated. This | In addition, the reliability of DNA analysis results from a low quality DNA data would be investigated. This can be approached by degrading several complete DNA data samples into different levels and conduct sets of genetic test on them. The change of test results will be observed and discussed. | ||
==Background== | ==Background== | ||
| Line 20: | Line 20: | ||
===DNA=== | ===DNA=== | ||
DNA is the hereditary material which stores the genetic information in humans | DNA is the hereditary material which stores the genetic information in humans. | ||
There are two types of DNA in human beings, one is known as nuclear DNA which is | There are two types of DNA in human beings, one is known as nuclear DNA which is | ||
located in cell nucleus and another type is mitochondrial DNA which is located in the | located in cell nucleus and another type is mitochondrial DNA which is located in the | ||
| Line 46: | Line 46: | ||
[[File:chromosome1.png|thumb|300px|Figure 3: Chromosome structure]] [[File:chromosome2.png|thumb|300px|Figure 4: 23 pairs of chromosomes in human]] | [[File:chromosome1.png|thumb|300px|Figure 3: Chromosome structure]] [[File:chromosome2.png|thumb|300px|Figure 4: 23 pairs of chromosomes in human]] | ||
Chromosome is an integrated package of DNA molecules. It has thread-like | Chromosome is an integrated package of DNA molecules. It has thread-like | ||
structures, and DNA molecules are coiled up around hi stones proteins to form the | |||
structure | structure. There are 23 pairs of chromosomes in human body's cell, which is 46 | ||
chromosomes in total. 22 pairs are called autosomes which are common for both | chromosomes in total. 22 pairs are called autosomes which are common for both | ||
males and females and the last 23rd pair is sex chromosomes which differ males and | males and females and the last 23rd pair is sex chromosomes which differ males and | ||
females. In this project, the DNA data analysis would only focus on autosomes | females. In this project, the DNA data analysis would only focus on autosomes. | ||
===SNP=== | ===SNP=== | ||
Single nucleotide | Single nucleotide polymorphism (SNP) is a genetic variation | ||
among human beings | among human beings. Each SNP represents a difference in a nucleotide which is | ||
a single DNA molecule | a single DNA molecule. For instance, one SNP may replace a nucleotide of base | ||
guanine (G) with cytosine (C). These SNPs can be found nearly once in every 1,000 | guanine (G) with cytosine (C). These SNPs can be found nearly once in every 1,000 | ||
nucleotides on average in a person's DNA. Most SNPs do not effect health of owner. | |||
However, some of these variations may | However, some of these variations may associate with diseases. | ||
===DNA reference file=== | ===DNA reference file=== | ||
A DNA reference file stores a group of SNPs data of owner's DNA. The format of DNA reference files | A DNA reference file stores a group of SNPs data of owner's DNA. The format of DNA reference files used in this project are of the same format which is 23andMe company's file, where 23andMe is a company that attended to provide personal genetic information for customers by using advanced genetic analysis techniques and web-based interactive tools. A screenshot of a sample file is shown below. | ||
[[File:Dna_ref.png|thumb|center|300px|Figure 5: Sample DNA file from 23andMe]] | [[File:Dna_ref.png|thumb|center|300px|Figure 5: Sample DNA file from 23andMe]] | ||
As shown in the figure, there are 4 columns rsid, | As shown in the figure, there are 4 columns in the DNA reference file: rsid, chromosome, position and genotype. The rsid is a unique id used to identify a specific SNP. | ||
The format of rsid starts with “rs” and followed by a number (eg. rs123456). These rsids are commonly used by researchers and databases. There is another special format of rsid that starts with “i” and followed by a number (eg. i123456). This “i” format is used internally by 23andMe to identify the unknown SNP and | The format of rsid starts with “rs” and followed by a number (eg. rs123456). These rsids are commonly used by researchers and databases. There is another special format of rsid that starts with “i” and followed by a number (eg. i123456). This “i” format is used internally by 23andMe to identify the unknown SNP and cannot be used in public database. The second column chromosome identify which chromosome the SNP belongs to (1st to 22nd chromosome). The third column, position, indicates positions of SNPs in owner's DNA sequence. The last column, genotype, is the column for base pairs of variants (A, T, G, C). Note that there are some cases where the genotype result for some SNPs are not able be provided and “--” would be displayed in genotype column. It is important to note that only the SNPs with identified base pairs can be used for DNA analysis. | ||
==Task 1: Testing with Somerton Man’s DNA reference file== | ==Task 1: Testing with Somerton Man’s DNA reference file== | ||
===Aims=== | ===Aims=== | ||
The aim of this task is to have a basic understanding of the DNA reference file and DNA analysis techniques. The project | The aim of this task is to have a basic understanding of the DNA reference file and DNA analysis techniques. The project provides a DNA reference file of the Somerton Man which is a corrupted DNA data. A screenshot of the file is shown in figure 6. | ||
Man which is a corrupted DNA data. A | |||
[[File:sm_dna.png|thumb|center|300px|Figure 6: Screenshot of Somerton Man's DNA reference file]] | [[File:sm_dna.png|thumb|center|300px|Figure 6: Screenshot of Somerton Man's DNA reference file]] | ||
The first goal of this task is to evaluate the quality of the file including counting the total amount of SNPs and the amount of available SNPs. | The first goal of this task is to evaluate the quality of the file including counting the total amount of SNPs and the amount of available (non-empty) SNPs. The second goal is to conduct some DNA analysis on the DNA reference file. | ||
===Methods=== | ===Methods=== | ||
To approach the first goal of this | To approach the first goal of this project, the team developed a program which provide functions for counting total amount of SNPs, amount of available SNPs (SNPs that do not have genotype of “--”), and determine the percentages of available SNPs for chromosome 1 to 22 of the Somerton Man’s DNA raw data. The program was developed using C++ language. | ||
A website called GEDmatch was used for conducting DNA analysis. GEDmatch is a website that has an open data personal genomics database and provide tools for DNA and genealogy research. The site become well known after law enforcement in California use it for the Golden State Killer case and are commonly used by all law enforcement in United States. The Somerton Man’s DNA reference file was uploaded to the website and several DNA analysis tools provided on the website was used. | |||
===Results and discussion=== | ===Results and discussion=== | ||
The counting outputs of Somerton man's DNA data is presented in figure 7. As the figure shown, there are more than 0.6 million SNPs in the files, but only about 2% of them have determined base pairs. In DNA analysis, only the SNPs with available base pairs can be used and most genetic analysis technique would require a certain amount of available SNPs. | The counting outputs of Somerton man's DNA data is presented in figure 7. As the figure shown, there are more than 0.6 million SNPs in the files, but only about 2% of them have determined base pairs. In DNA analysis, only the SNPs with available base pairs can be used and most genetic analysis technique would require a certain amount of available SNPs. | ||
[[File:SM_Counting.png|thumb|center|300px|Figure 7: SNPs counting results of Somerton man DNA file]] | [[File:SM_Counting.png|thumb|center|300px|Figure 7: SNPs counting results of Somerton man DNA file]] | ||
Then the Somerton Man's DNA reference file was uploaded to GEDmatch | Then the Somerton Man's DNA reference file was uploaded to GEDmatch to be used on the one-to-many tool. One-to-many tool is the main service provided by GEDmatch. When a DNA raw data reference file is updated, it will be stored as a kit in GEDmatch database, and be compared with other kits in the public database. After the matching process has finished, the one-to-many tool can show how many kits in database match with the kit that the user has uploaded. Unfortunately the website rejects to process the Somerton Man's data to use the one-to-many tool since the file did not meet the minimum requirements of 2000 SNPs for each chromosome. | ||
[[File:SNP_low.png|thumb|600px|center|Figure 8: DNA kit not accepted due to low amount of SNPs]] | [[File:SNP_low.png|thumb|600px|center|Figure 8: DNA kit not accepted due to low amount of SNPs]] | ||
===Conclusion=== | ===Conclusion=== | ||
The quality of Somerton Man's DNA reference file is lower than expected. Only about 2% of 613905 SNPs in the files are available for use. Such low quality DNA file is not accepted by GEDmatch to conduct DNA match examination. In order to satisfy the minimum requirements of GEDmatch, a data recovery work would be required which | The quality of Somerton Man's DNA reference file is lower than expected. Only about 2% of 613905 SNPs in the files are available for use. Such low quality DNA file is not accepted by GEDmatch to conduct DNA match examination. In order to satisfy the minimum requirements of GEDmatch, a data recovery work would be required which will be introduced in the next task. | ||
==Task 2: Artificially recover DNA file== | ==Task 2: Artificially recover DNA file== | ||
===Aims=== | ===Aims=== | ||
In this task, the project group aims to artificially recover Somerton Man's DNA file to satisfy the basic SNPs amount requirements (2000 SNPs for each chromosome) of GEDmatch's one-to-many tool and find out how many people is | In this task, the project group aims to artificially recover Somerton Man's DNA file to satisfy the basic SNPs amount requirements (2000 SNPs for each chromosome) of GEDmatch's one-to-many tool and find out how many people is related to Somerton Man's DNA kit. | ||
===Methods=== | ===Methods=== | ||
The recovery works | The recovery works was done by developing multiple programs using C++. In general, the recovery work is to replace a fixed amount of empty SNPs which is 2000 SNPs for each chromosome with available SNPs. Several simple recovery algorithms were implemented. The first algorithm is called random algorithm which is to replace empty SNPs with random base pairs in genotype. The second algorithm used was by replacing empty genotype with homozygous pairs (AA, GG, TT, CC) which resulted in 4 new algorithms. | ||
In addition, if there | In addition, if there was no DNA kit that matches with Somerton Man' DNA in the database, recovering more empty SNP's of the Somerton Man's DNA could be a back up plan. With the recovery algorithm introduced before, the project team can recover more SNPs in Somerton Man's DNA reference file and try to use the one-to-many tool on those kits. | ||
===Results and discussion=== | ===Results and discussion=== | ||
With the developed program, multiple artificial DNA kits which have 2000 SNPs in each chromosome were created. Unfortunately, all of these DNA kits have 0 matches with other DNA in the public database which means these artificial DNA kits | With the developed program, multiple artificial DNA kits which have 2000 SNPs in each chromosome were created. Unfortunately, all of these DNA kits have 0 matches with other DNA in the public database which means these artificial DNA kits do not relate to any kit in the GEDmatch database. | ||
[[File:zero_match.png|thumb|600px|center|Figure 9: match results of artificial DNA(replace empty SNPs with random pairs to 2000 SNPs in each choromosone)]] | [[File:zero_match.png|thumb|600px|center|Figure 9: match results of artificial DNA(replace empty SNPs with random pairs to 2000 SNPs in each choromosone)]] | ||
Then kits with more amount of empty SNPs were replaced with homozygous pairs or random pairs were created, but none of these files could find relative DNA kits in the database. Even the DNA kits with all empty SNPs recovered could find a matched DNA result. It is important to note that all 5 recovery strategies were all implemented. | Then kits with more amount of empty SNPs were replaced with homozygous pairs or random pairs were created, but none of these files could find relative DNA kits in the database. Even the DNA kits with all empty SNPs recovered could find a matched DNA result. It is important to note that all 5 recovery strategies were all implemented. | ||
As GEDmatch is the most commonly used DNA database in public, it | As GEDmatch is the most commonly used DNA database in public, it contains a huge amount of DNA kits in its database. As the website shown, the total number of kits managed by GEDmatch database is 1363427157376. Therefore, the chance that no DNA kit in the database is related to Somerton Man is nearly impossible. This means that the quality of Somerton Man's DNA file is too low to be used on the one-to-many tool and implementing simple recovery algorithms are pointless. | ||
===Conclusion=== | ===Conclusion=== | ||
It is obvious that there is too many empty SNPs in Somerton Man's DNA reference file. | It is obvious that there is too many empty SNPs in Somerton Man's DNA reference file. The recovery algorithms introduced were too simple and cannot help to increase the chance of finding Somerton Man's relatives in the GEDmatch database. With DNA data that only contains approximate 2% available SNPs, it is nearly impossible to find any possible related DNA kits to Somerton Man. | ||
==Task 3: Investigation on ethnicity== | ==Task 3: Investigation on ethnicity== | ||
===Aims=== | ===Aims=== | ||
The first aim of this task is | The first aim of this task is to investigate the ethnicity of the Somerton Man. As described in previous section, the quality of Somerton Man's DNA is low, therefore the second aim is to study the reliability of low quality DNA's ethnicity examination results. | ||
===Methods=== | ===Methods=== | ||
An ethnicity tool called Eurogenes Ad-Mix Utilities was used. This tool was provided by GEDmatch and can generate a report of ethnicity proportions to the given DNA kit. Eurogenes K13 model is selected as the 'calculator' model. This model calculates and gives results of the ethnicity proportion in 13 different global regions as shown in Figure 10, and this mode is primarily for European background people since it provides more sub-continental regions for Europe. The Somerton Man's DNA was selected as input kit of the utility and the ethnicity report was generated. | |||
[[File:ethnicity_sample.png|thumb|300px|center|Figure 10: A sample report of Eurogenes Ad-Mix Utilities]] | [[File:ethnicity_sample.png|thumb|300px|center|Figure 10: A sample report of Eurogenes Ad-Mix Utilities]] | ||
In addition, to investigate the reliability of a low quality DNA data file's ethnicity report, several complete DNA samples | |||
In order to provide stronger evidence to prove whether the low quality DNA file's ethnicity report is reliable or not, different degradation algorithms | In addition, to investigate the reliability of a low quality DNA data file's ethnicity report, several complete DNA samples was analysed. The project ordered 2 sets of complete DNA reference data from 23andMe which provide same format as Somerton Man's file. A program was developed that allows the user to degrade the selected DNA file into different levels of DNA data. This program was also developed using C++. The project team degraded each complete DNA sample files into 9 files by removing 10% SNPs, 20% SNPs and then step by step to 90% SNPs. An extra file which contains only the SNPs with same rsids in Somerton Man's DNA file was created and was named as degraded_DNA for each set of complete DNA sample data. These files were then uploaded to GEDmatch and the same ethnicity research was conducted as what has been done on Somerton man's DNA raw data. All ethnicity reports were recorded, and the change of how the ethnicity proportion changes was also observed. | ||
In order to provide stronger evidence to prove whether the low quality DNA file's ethnicity report is reliable or not, different degradation algorithms were introduced. The first strategy was that for every 10 SNPs, the first n% SNPs were removed where n% is the percentage of SNPs we would like to remove. The next algorithm performed was the opposite of the first algorithm. This algorithm removed the last n% SNPs for every 10 SNPs, where n% is the percentage of SNPs we would like to remove. The third and fourth methods were to remove the first and last n% of SNPs for each chromosome, where n% is the percentage of SNPs we would like to remove. | |||
===Results and discussion=== | ===Results and discussion=== | ||
The ethnicity report of Somerton Man's DNA are shown in | The ethnicity report of Somerton Man's DNA are shown in Figure 11. As the shown in the pie chart, the first 2 major regions are North Atlantic region which contributes up to 36.21% of the chart, and Baltic region which is 20.44%. | ||
[[File:ethnicity_SM.png|thumb|400px|center|Figure 11: Ethnicity report of Somerton Man's DNA]] | [[File:ethnicity_SM.png|thumb|400px|center|Figure 11: Ethnicity report of Somerton Man's DNA]] | ||
According to the population averages table[15] for Eurogenes K13 model provided by the developer Davidski (Polako), both Baltic and North Atlantic regions are in Europe. | According to the population averages table[15] for Eurogenes K13 model provided by the developer Davidski (Polako), both Baltic and North Atlantic regions are in Europe. Figure 12 is a map that indicates the areas of Baltic region and Figure 13 shows North Atlantic region. | ||
<div style="text-align:center;"><ul> | <div style="text-align:center;"><ul> | ||
<li style="display: inline-block;"> [[File:Baltic.png|right|thumb|300px|Figure 12: Map of Baltic region]] </li> | <li style="display: inline-block;"> [[File:Baltic.png|right|thumb|300px|Figure 12: Map of Baltic region]] </li> | ||
<li style="display: inline-block;"> [[File:North_atlantic.png|right|thumb|323px|Figure 13: Map of North Atlantic region]] </li> | <li style="display: inline-block;"> [[File:North_atlantic.png|right|thumb|323px|Figure 13: Map of North Atlantic region]] </li> | ||
</ul></div> | </ul></div> | ||
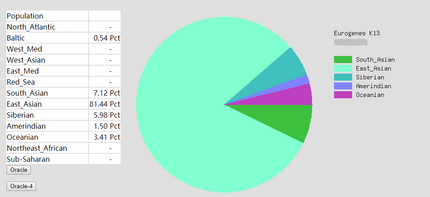
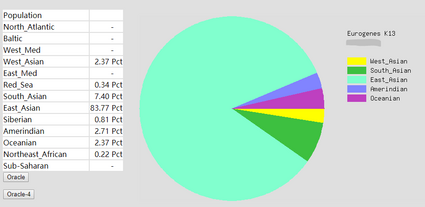
To prove the ethnicity report created | To prove the ethnicity report created was reliable, 2 complete DNA files were gained and were degraded to the same level of Somerton Man's DNA which is 2% SNPs remaining in the file. Sample DNA reference file 1 contained 613967 SNPs and 96.41% of them were not empty, and DNA reference file 2 has 614009 SNPs and 97.68% of them were available for use. The ethnicity reports of 2 complete sample DNA files are presented in figure 16 and 17. Also, ethnicity reports of degraded_DNA files for each complete DNA are shown in figure 14 and 15. According to the ethnicity reports shown in those figures, the proportion of the largest and second largest ethnicity regions of sample DNA file 1 have changed to 83.13% to 78.66% and 14.82% to 18.14% after degradation process. The first major region proportion has reduced 4.64% and the second region proportion has increased for 3.32%. The degradation process affected the proportion of each ethnicity region for DNA sample 1, but the change is not much and the first and second regions are still the largest 2 regions in the pie chart. Similar phenomenon can be discovered when comparing ethnicity reports of DNA sample 2. The largest ethnicity regions has grown for 2.33% from 81.44% to 83.77%, and the second largest region increased 0.28% from 7.12% to 7.40%. These changes shows that the proportion of major ethnicity regions would not change greatly when a complete human DNA file is degraded to a level of 2% SNPs remaining. | ||
<div style="text-align:center;"><ul> | <div style="text-align:center;"><ul> | ||
<li style="display: inline-block;"> [[File:s1_eth.png|right|thumb|410px|Figure 14: Ethnicity reports of sample DNA file 1]] </li> | <li style="display: inline-block;"> [[File:s1_eth.png|right|thumb|410px|Figure 14: Ethnicity reports of sample DNA file 1]] </li> | ||
| Line 126: | Line 136: | ||
<li style="display: inline-block;"> [[File:degrade_s2_eth.png|right|thumb|425px|Figure 17: Ethnicity reports of sample DNA file 2 after degradation]] </li> | <li style="display: inline-block;"> [[File:degrade_s2_eth.png|right|thumb|425px|Figure 17: Ethnicity reports of sample DNA file 2 after degradation]] </li> | ||
</ul></div> | </ul></div> | ||
To provide more | To provide more evidence to prove this theory, several degradation algorithms introduced in section 5.2 have been applied and changes of ethnicity proportions during different degradation processes have been observed and recorded. 2 sample DNA reference files were degraded into 9 files at different levels from 90% to 10% SNPs remaining. The proportion of first 2 largest ethnicity regions of each degraded files have been plotted on line graphs. Figure 18 is the line graph that shows how the means of ethnicity proportions change via the degradation process with standard error provided. As the graph shown, each region proportion fluctuate at a certain level. For instance the percentage of first region of sample 1 fluctuate at around 83% which is a close value to the original proportion 81.44%. However, error bars or standard errors of each region become larger, as more SNPs are removed, which indicate that as more SNPs being removed, the proportions presented in ethnicity reports become less accurate. But in another case, the highest standard error for first and second region proportions of sample 1 and 2 are 1.32%, 1.41%, 1.33% and 1.03%. None of these standard errors exceed 1.5% which can be seen as an acceptable errors. Therefore the project concludes that when a large amount of SNPs are removed from a set of DNA data, the ethnicity report generated from the DNA data would be influenced, but the results are still acceptable to identify the owner's ethnicity. | ||
[[File:eth_line_graph.png|thumb|600px|center|Figure 18: Line graph of means of ethnicity proportions vs degradation level]] | [[File:eth_line_graph.png|thumb|600px|center|Figure 18: Line graph of means of ethnicity proportions vs degradation level]] | ||
===Conclusion=== | ===Conclusion=== | ||
According to the observation of ethnicity change during the degradation process, as more amount of SNPs are removed from a complete human DNA reference file, the result of ethnicity report would be less | According to the observation of ethnicity change during the degradation process, as more amount of SNPs are removed from a complete human DNA reference file, the result of ethnicity report would be less accurate but the largest and second largest ethnicity regions in the report are still reliable. Therefore the top two major ethnicity of the Somerton Man are North Atlantic and Baltic, where these two regions are mostly around Europe. | ||
==Genetic diseases search== | ==Genetic diseases search== | ||
===Aims=== | ===Aims=== | ||
During this task, the team | During this task, the team focused on searching clinical effects of each available SNP and identify any possible genetic disease or physical characteristics that Somerton Man could have. | ||
===Methods=== | ===Methods=== | ||
To search the clinical effects of SNPs, the team | To search the clinical effects of SNPs, the team developed a data mining program that collects information in SNP database. Python language was used for development since it is convenient for web development. The SNP database the project selected to use was dbSNP which is the largest database for nucleotide variations in the world, and is managed by the National Center for Biotechnology Information (NCBI). Figure 6.1 shows the information provided by dbSNP. The project team collected the clinical significance related to each rsid in Somerton Man's file. | ||
[[File:dbSNP.png|thumb|600px|center|Figure 19:information of SNP rs12913832]] | [[File:dbSNP.png|thumb|600px|center|Figure 19:information of SNP rs12913832]] | ||
The program | |||
The program extracted every non-empty SNP in Somerton Man's DNA reference file. With the API provided by dbSNP, connection to dbSNP was established and each rsid of the extracted SNP was sent. When the connection was successfully set up, dbSNP sent back the information of corresponding SNP in JSON format. The data sent back was analysed and clinical information such as genetic disease name associated with the SNPs was recorded. | |||
===Result and discussion=== | ===Result and discussion=== | ||
With the support of data mining program, 613905 SNPs were searched in the database and 574 diseases were found. Figure 19 shows part of the genetic diseases outputs. As the figure shown, the program | With the support of data mining program, 613905 SNPs were searched in the database and 574 diseases were found. Figure 19 shows part of the genetic diseases outputs. As the figure shown, the program recorded the rsid of SNP that the disease belonged to in rsid column. dbSNP provides only a brief description of the clinical effects. More details are linked to another database called ClinVar which is a freely accessible, public database that provide medical reports of the relationships among human variants and phenotypes [12]. Therefore ClinVar Accession column is introduced to collect the ID of the recorded disease. This ID linked to the Clinvar database and allow the user to find a detailed medical report about the disease. The diseases names are recorded in disease name column. It is necessary to indicate that there are multiple diseases named with 'not specified' or 'not provided' which requires the user to find a detailed description of the disease in Clinvar. Unfortunately, none of diseases in the results relates or corresponds to Somerton Man's known characteristics. | ||
[[File:disease_list.png|thumb|600px|center|Figure 19: Outputs of data mining program]] | [[File:disease_list.png|thumb|600px|center|Figure 19: Outputs of data mining program]] | ||
| Line 171: | Line 184: | ||
==Conclusions== | ==Conclusions== | ||
The Somerton Man's DNA reference file provided to the project contain 613905 SNPs, but only 2.08% of SNPs | The Somerton Man's DNA reference file provided to the project contain 613905 SNPs, but only 2.08% of SNPs that were not empty and were available for DNA analysis. With such low portion of available SNP, limited DNA analysis techniques can be conducted on the file. | ||
Unfortunately, there | Unfortunately, there was no DNA kit that matched with Somerton Man's DNA kit found in GEDmatch database. According to the result of task 2 and task 4, the degradation process would have huge effect on the match results of a human DNA data. And it is impossible to recover Somerton Man's DNA by implementing simple recovery methods such as replacing empty SNPs with random base pairs or homozygous pairs. But if some reliable recovery strategies were introduced which have not been determined yet and allow Somerton Man's DNA to be recovered to more than 60% SNPs, then his relatives may be discovered. | ||
Moreover, | Moreover, the result from task 4 shows that the Somerton Man originated from Europe. To be specific, his ethnicity is about 36.21% North Atlantic and 20.44% Baltic. | ||
As for the genetic disease, 574 diseases were found, but there | As for the genetic disease, 574 diseases were found, but there was no disease found that relates to his known appearance. | ||
So far, that is what the project can | So far, that is what the project can find in regards to the Somerton Man's DNA data. There was no clear clue that can lead to his identity. Who the Somerton Man is will still be a mystery. | ||
==Future Work== | ==Future Work== | ||
| Line 183: | Line 196: | ||
==Reference== | ==Reference== | ||
[1]Bineth, J, "Somerton Man: One of Australia's most baffling cold cases could be a step closer to being solved" This Is About, 13 December 2017. [online] Available at: https://www.abc.net.au/news/2017-12-14/somerton-man-cold-case-could-be- one-step-closer-to-solved/9245512 [Accessed 1 Jun. 2019]. | [1] Bineth, J, "Somerton Man: One of Australia's most baffling cold cases could be a step closer to being solved" This Is About, 13 December 2017. [online] Available at: https://www.abc.net.au/news/2017-12-14/somerton-man-cold-case-could-be- one-step-closer-to-solved/9245512 [Accessed 1 Jun. 2019]. | ||
[2] U.S. National Library of Medicine, "What is DNA?",U.S. National Library of Medicine, May. 28, 2019. [online] Available at: https://ghr.nlm.nih.gov/primer/basics/dna [Accessed 2 Jun. 2019]. | |||
[ | [3] U.S. National Library of Medicine, "What is a chromosome?",U.S. National Library of Medicine, May. 28, 2019. [online] Available at: https://ghr.nlm.nih.gov/primer/basics/chromosome [Accessed 2 Jun. 2019]. | ||
[ | [4] U.S. National Library of Medicine, "How many chromosomes do people have?",U.S. National Library of Medicine, May. 28, 2019. [Online]. Available: https://ghr.nlm.nih.gov/primer/basics/howmanychromosomes. [Accessed: 02- Jun- 2019]. | ||
[ | [5] U.S. National Library of Medicine, "What are single nucleotide polymorphisms (SNPs)?",U.S. National Library of Medicine, May. 28, 2019. [Online]. Available: https://ghr.nlm.nih.gov/primer/genomicresearch/snp. [Accessed: 02- Jun- 2019]. | ||
[ | [6] G. Shaw. “Polymorphism and Single nucleotide polymorphisms (SNPs)” Science Made Simple, Vol. 112, pp.664-665 2013. | ||
[ | [7] “DEAD MAN FOUND LYING ON SOMERTON BEACH” The News, December 1, 1948, p. 1 [online]. Available: https://trove.nla.gov.au/newspaper/article/129897161. [Accessed: 03- Jun- 2019]. | ||
[ | [8] “Cryptic Note On Body” The News, June 6, 1949, p. 1 [online]. Available: https://trove.nla.gov.au/newspaper/article/36371152. [Accessed: 03- Jun- 2019]. | ||
[ | [9] "Raw Data Technical Details", 23andMe, 2019. [Online]. Available: https://customercare.23andme.com/hc/en-us/articles/115004459928-Raw-Data- Technical-Details. [Accessed: 04- Jun- 2019]. | ||
[ | [10] S. Zhang, "The Coming Wave of Murders Solved by Genealogy", The Atlantic, 2019. [Online]. Available: https://www.theatlantic.com/science/archive/2018/05/the-coming-wave-of- murders-solved-by-genealogy/560750/. [Accessed: 04- Jun- 2019]. | ||
[ | [11] "General Information about dbSNP as a Database Resource", Center for Biotechnology Information (US), 2005. [Online]. Available: https://www.ncbi.nlm.nih.gov/books/NBK44469/. [Accessed: 06- Jun- 2019]. | ||
[ | [12] Landrum, M., Lee, J., Riley, G., Jang, W., Rubinstein, W., Church, D. and Maglott, D. (2013). ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Research, 42(D1), pp.D980-D985. | ||
[ | [13] Chick, H. (2017). Finally! A Gedmatch Admixture Guide!. [Blog] genealogical musings. Available at: https://genealogical-musings.blogspot.com/2017/04/finally-gedmatch-admixture-guide.html [Accessed 29- Oct- 2019]. | ||
[ | [14] Chen, J. and Seroka, A. (2018). Cipher cracking Final Report/Thesis 2018. [online] Eleceng.adelaide.edu.au. Available at: https://www.eleceng.adelaide.edu.au/personal/dabbott/wiki/index.php/Final_Report/Thesis_2018 [Accessed 1 Nov. 2019]. | ||
[ | [15] Davidski. (2019). K13_population_averages. [online] Available at: https://docs.google.com/spreadsheets/d/1Oz6P5-SVEJciPX1TciGe-zoqA5JtOGIMG7nh-rCOj0c/edit#gid=804264822 [Accessed 1 Nov. 2019]. | ||
[ | [16] Inside Story, presented by Stuart Littlemore, ABC TV, screened at 8 pm, Thursday, 24th August, 1978 | ||
==Appendix A: Codes and tables used in the project== | |||
==Appendix A: Codes used in the project== | *[https://github.com/ratherto/Who-killed-the-Somerton-Man-2019 Code and tables used for the project] | ||
* | |||
==Appendix B: Tables used for the graph== | ==Appendix B: Tables used for the graph== | ||
*ethnicity line graph:[[File:ethnicity changes. | *ethnicity line graph:[[File:ethnicity changes.zip]] | ||
*false positives and false negatives test: [[File:false. | *false positives and false negatives test: [[File:false.zip]] | ||
Latest revision as of 11:00, 7 November 2019
Abstract[edit]
The Somerton Man's case is one of the most mysterious case in South Australia until today. An unknown man was murdered on the Somerton beach, and the identification of the victim is still a mystery. The project aims to investigate the identification of the Somerton man by means of studying his DNA data provided. Unfortunately, the DNA data was corrupted and has a high drop rate, therefore the project team was required to use different strategies and techniques to recover and analyse the DNA, then try to find out any possible characteristics of the Somerton Man.
To approach the goals of the project, the team firstly evaluates the DNA data and try to conduct DNA analysis via different genetic services. In addition, a degradation process of complete DNA data was done. By degrading several complete DNA files, we can observe how degradation could affect DNA analysis results.
Introduction[edit]
Motivation[edit]
The main topic of the project is human identification using software programming and genetic analysis techniques. The project conducts a study on investigating the identification of the victim, Somerton Man, which is one of the most mysterious cases in South Australia until today. On December 1st 1948, a well-dressed male was found dead on Somerton Beach in Adelaide. He was clean-shaven, well dressed in a suit and no belongings of him that could prove his identity. Later the man was called as the Somerton Man. The figure below shows the look of the Somerton Man.
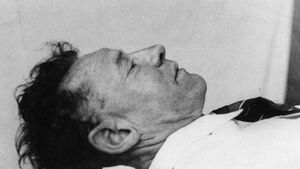
After more than half century, the identification of the Somerton Man is still unknown. With the Somerton Man's DNA data extracted from his hair, the project team may be able to conduct several DNA examinations and identify characteristics of the man. Unfortunately, the DNA data is corrupted due to degradation of the hair, but the team of the project will try the best to investigate the data with modern techniques. In modern society, human identification techniques is useful in multiple aspects, such as criminal investigation or seeking relatives. The most current identification techniques would require high quality DNA samples, but the project focus on investigating identification techniques based on low quality genetic data. Also the project concentrates on using engineering methods and problem solving skills to improve the identification techniques.
Objectives[edit]
The aim of the project is to investigate the identification of the Somerton Man. To be more specific, the group is aiming to identify any possible relatives, physical characteristics, genetic diseases or ethnicity of the Somerton Man. To achieve these goals, the team used software and genetic analysis techniques to work on the Somerton Man's DNA data. In addition, the reliability of DNA analysis results from a low quality DNA data would be investigated. This can be approached by degrading several complete DNA data samples into different levels and conduct sets of genetic test on them. The change of test results will be observed and discussed.
Background[edit]
DNA[edit]
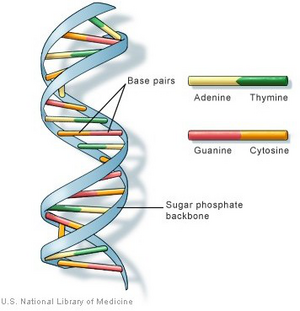
DNA is the hereditary material which stores the genetic information in humans. There are two types of DNA in human beings, one is known as nuclear DNA which is located in cell nucleus and another type is mitochondrial DNA which is located in the mitochondria. This project only focuses on the analysis of nuclear DNA. DNA stores genetic information as a sequence built up with four types of nitrogen bases which are adenine (A), guanine (G), cytosine (C), and thymine (T) [2]. Also, a sugar molecule and a phosphate molecule are attached to each nitrogen base to form a molecule called nucleotide. The bases would pair up (A with T and C with G) and multiple nucleotides are placed in two strands to form a double helix which looks like a spiral [2]. In general, a DNA is a genetic sequence formed by multiple base pairs. The genetic instructions of building and maintaining an organism are obtained from the order of these base pairs [2]. There are about 3 billion bases in human DNA, in which more than 99% of the bases are common in all human beings, and the physiological differences among people depends on these 1% DNA.
Chromosome[edit]
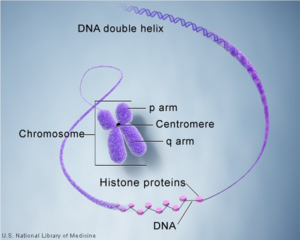
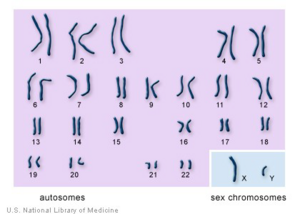
Chromosome is an integrated package of DNA molecules. It has thread-like structures, and DNA molecules are coiled up around hi stones proteins to form the structure. There are 23 pairs of chromosomes in human body's cell, which is 46 chromosomes in total. 22 pairs are called autosomes which are common for both males and females and the last 23rd pair is sex chromosomes which differ males and females. In this project, the DNA data analysis would only focus on autosomes.
SNP[edit]
Single nucleotide polymorphism (SNP) is a genetic variation among human beings. Each SNP represents a difference in a nucleotide which is a single DNA molecule. For instance, one SNP may replace a nucleotide of base guanine (G) with cytosine (C). These SNPs can be found nearly once in every 1,000 nucleotides on average in a person's DNA. Most SNPs do not effect health of owner. However, some of these variations may associate with diseases.
DNA reference file[edit]
A DNA reference file stores a group of SNPs data of owner's DNA. The format of DNA reference files used in this project are of the same format which is 23andMe company's file, where 23andMe is a company that attended to provide personal genetic information for customers by using advanced genetic analysis techniques and web-based interactive tools. A screenshot of a sample file is shown below.
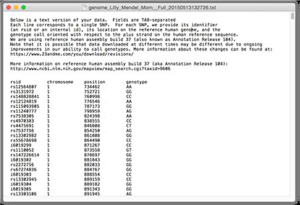
As shown in the figure, there are 4 columns in the DNA reference file: rsid, chromosome, position and genotype. The rsid is a unique id used to identify a specific SNP. The format of rsid starts with “rs” and followed by a number (eg. rs123456). These rsids are commonly used by researchers and databases. There is another special format of rsid that starts with “i” and followed by a number (eg. i123456). This “i” format is used internally by 23andMe to identify the unknown SNP and cannot be used in public database. The second column chromosome identify which chromosome the SNP belongs to (1st to 22nd chromosome). The third column, position, indicates positions of SNPs in owner's DNA sequence. The last column, genotype, is the column for base pairs of variants (A, T, G, C). Note that there are some cases where the genotype result for some SNPs are not able be provided and “--” would be displayed in genotype column. It is important to note that only the SNPs with identified base pairs can be used for DNA analysis.
Task 1: Testing with Somerton Man’s DNA reference file[edit]
Aims[edit]
The aim of this task is to have a basic understanding of the DNA reference file and DNA analysis techniques. The project provides a DNA reference file of the Somerton Man which is a corrupted DNA data. A screenshot of the file is shown in figure 6.
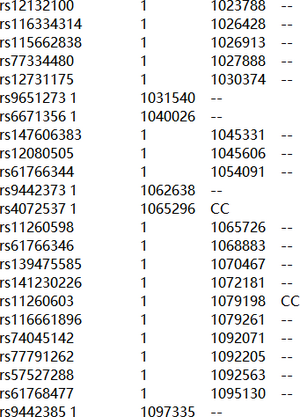
The first goal of this task is to evaluate the quality of the file including counting the total amount of SNPs and the amount of available (non-empty) SNPs. The second goal is to conduct some DNA analysis on the DNA reference file.
Methods[edit]
To approach the first goal of this project, the team developed a program which provide functions for counting total amount of SNPs, amount of available SNPs (SNPs that do not have genotype of “--”), and determine the percentages of available SNPs for chromosome 1 to 22 of the Somerton Man’s DNA raw data. The program was developed using C++ language. A website called GEDmatch was used for conducting DNA analysis. GEDmatch is a website that has an open data personal genomics database and provide tools for DNA and genealogy research. The site become well known after law enforcement in California use it for the Golden State Killer case and are commonly used by all law enforcement in United States. The Somerton Man’s DNA reference file was uploaded to the website and several DNA analysis tools provided on the website was used.
Results and discussion[edit]
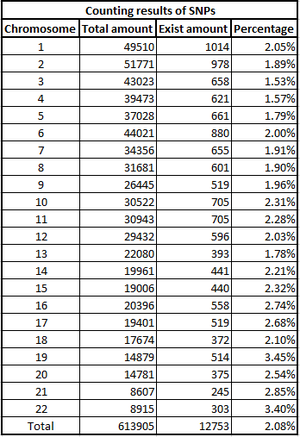
The counting outputs of Somerton man's DNA data is presented in figure 7. As the figure shown, there are more than 0.6 million SNPs in the files, but only about 2% of them have determined base pairs. In DNA analysis, only the SNPs with available base pairs can be used and most genetic analysis technique would require a certain amount of available SNPs.
Then the Somerton Man's DNA reference file was uploaded to GEDmatch to be used on the one-to-many tool. One-to-many tool is the main service provided by GEDmatch. When a DNA raw data reference file is updated, it will be stored as a kit in GEDmatch database, and be compared with other kits in the public database. After the matching process has finished, the one-to-many tool can show how many kits in database match with the kit that the user has uploaded. Unfortunately the website rejects to process the Somerton Man's data to use the one-to-many tool since the file did not meet the minimum requirements of 2000 SNPs for each chromosome.
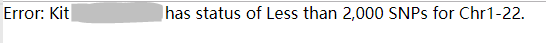
Conclusion[edit]
The quality of Somerton Man's DNA reference file is lower than expected. Only about 2% of 613905 SNPs in the files are available for use. Such low quality DNA file is not accepted by GEDmatch to conduct DNA match examination. In order to satisfy the minimum requirements of GEDmatch, a data recovery work would be required which will be introduced in the next task.
Task 2: Artificially recover DNA file[edit]
Aims[edit]
In this task, the project group aims to artificially recover Somerton Man's DNA file to satisfy the basic SNPs amount requirements (2000 SNPs for each chromosome) of GEDmatch's one-to-many tool and find out how many people is related to Somerton Man's DNA kit.
Methods[edit]
The recovery works was done by developing multiple programs using C++. In general, the recovery work is to replace a fixed amount of empty SNPs which is 2000 SNPs for each chromosome with available SNPs. Several simple recovery algorithms were implemented. The first algorithm is called random algorithm which is to replace empty SNPs with random base pairs in genotype. The second algorithm used was by replacing empty genotype with homozygous pairs (AA, GG, TT, CC) which resulted in 4 new algorithms. In addition, if there was no DNA kit that matches with Somerton Man' DNA in the database, recovering more empty SNP's of the Somerton Man's DNA could be a back up plan. With the recovery algorithm introduced before, the project team can recover more SNPs in Somerton Man's DNA reference file and try to use the one-to-many tool on those kits.
Results and discussion[edit]
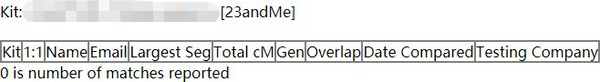
With the developed program, multiple artificial DNA kits which have 2000 SNPs in each chromosome were created. Unfortunately, all of these DNA kits have 0 matches with other DNA in the public database which means these artificial DNA kits do not relate to any kit in the GEDmatch database.
Then kits with more amount of empty SNPs were replaced with homozygous pairs or random pairs were created, but none of these files could find relative DNA kits in the database. Even the DNA kits with all empty SNPs recovered could find a matched DNA result. It is important to note that all 5 recovery strategies were all implemented. As GEDmatch is the most commonly used DNA database in public, it contains a huge amount of DNA kits in its database. As the website shown, the total number of kits managed by GEDmatch database is 1363427157376. Therefore, the chance that no DNA kit in the database is related to Somerton Man is nearly impossible. This means that the quality of Somerton Man's DNA file is too low to be used on the one-to-many tool and implementing simple recovery algorithms are pointless.
Conclusion[edit]
It is obvious that there is too many empty SNPs in Somerton Man's DNA reference file. The recovery algorithms introduced were too simple and cannot help to increase the chance of finding Somerton Man's relatives in the GEDmatch database. With DNA data that only contains approximate 2% available SNPs, it is nearly impossible to find any possible related DNA kits to Somerton Man.
Task 3: Investigation on ethnicity[edit]
Aims[edit]
The first aim of this task is to investigate the ethnicity of the Somerton Man. As described in previous section, the quality of Somerton Man's DNA is low, therefore the second aim is to study the reliability of low quality DNA's ethnicity examination results.
Methods[edit]
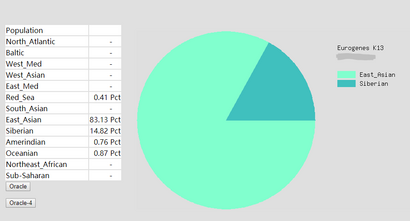
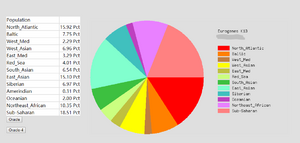
An ethnicity tool called Eurogenes Ad-Mix Utilities was used. This tool was provided by GEDmatch and can generate a report of ethnicity proportions to the given DNA kit. Eurogenes K13 model is selected as the 'calculator' model. This model calculates and gives results of the ethnicity proportion in 13 different global regions as shown in Figure 10, and this mode is primarily for European background people since it provides more sub-continental regions for Europe. The Somerton Man's DNA was selected as input kit of the utility and the ethnicity report was generated.
In addition, to investigate the reliability of a low quality DNA data file's ethnicity report, several complete DNA samples was analysed. The project ordered 2 sets of complete DNA reference data from 23andMe which provide same format as Somerton Man's file. A program was developed that allows the user to degrade the selected DNA file into different levels of DNA data. This program was also developed using C++. The project team degraded each complete DNA sample files into 9 files by removing 10% SNPs, 20% SNPs and then step by step to 90% SNPs. An extra file which contains only the SNPs with same rsids in Somerton Man's DNA file was created and was named as degraded_DNA for each set of complete DNA sample data. These files were then uploaded to GEDmatch and the same ethnicity research was conducted as what has been done on Somerton man's DNA raw data. All ethnicity reports were recorded, and the change of how the ethnicity proportion changes was also observed.
In order to provide stronger evidence to prove whether the low quality DNA file's ethnicity report is reliable or not, different degradation algorithms were introduced. The first strategy was that for every 10 SNPs, the first n% SNPs were removed where n% is the percentage of SNPs we would like to remove. The next algorithm performed was the opposite of the first algorithm. This algorithm removed the last n% SNPs for every 10 SNPs, where n% is the percentage of SNPs we would like to remove. The third and fourth methods were to remove the first and last n% of SNPs for each chromosome, where n% is the percentage of SNPs we would like to remove.
Results and discussion[edit]
The ethnicity report of Somerton Man's DNA are shown in Figure 11. As the shown in the pie chart, the first 2 major regions are North Atlantic region which contributes up to 36.21% of the chart, and Baltic region which is 20.44%.
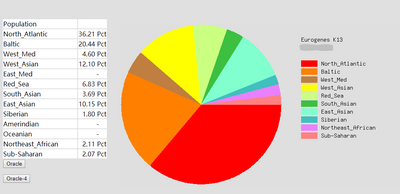
According to the population averages table[15] for Eurogenes K13 model provided by the developer Davidski (Polako), both Baltic and North Atlantic regions are in Europe. Figure 12 is a map that indicates the areas of Baltic region and Figure 13 shows North Atlantic region.
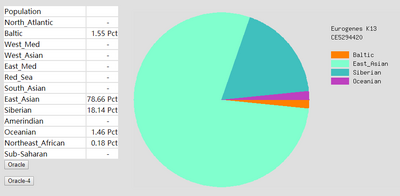
To prove the ethnicity report created was reliable, 2 complete DNA files were gained and were degraded to the same level of Somerton Man's DNA which is 2% SNPs remaining in the file. Sample DNA reference file 1 contained 613967 SNPs and 96.41% of them were not empty, and DNA reference file 2 has 614009 SNPs and 97.68% of them were available for use. The ethnicity reports of 2 complete sample DNA files are presented in figure 16 and 17. Also, ethnicity reports of degraded_DNA files for each complete DNA are shown in figure 14 and 15. According to the ethnicity reports shown in those figures, the proportion of the largest and second largest ethnicity regions of sample DNA file 1 have changed to 83.13% to 78.66% and 14.82% to 18.14% after degradation process. The first major region proportion has reduced 4.64% and the second region proportion has increased for 3.32%. The degradation process affected the proportion of each ethnicity region for DNA sample 1, but the change is not much and the first and second regions are still the largest 2 regions in the pie chart. Similar phenomenon can be discovered when comparing ethnicity reports of DNA sample 2. The largest ethnicity regions has grown for 2.33% from 81.44% to 83.77%, and the second largest region increased 0.28% from 7.12% to 7.40%. These changes shows that the proportion of major ethnicity regions would not change greatly when a complete human DNA file is degraded to a level of 2% SNPs remaining.
To provide more evidence to prove this theory, several degradation algorithms introduced in section 5.2 have been applied and changes of ethnicity proportions during different degradation processes have been observed and recorded. 2 sample DNA reference files were degraded into 9 files at different levels from 90% to 10% SNPs remaining. The proportion of first 2 largest ethnicity regions of each degraded files have been plotted on line graphs. Figure 18 is the line graph that shows how the means of ethnicity proportions change via the degradation process with standard error provided. As the graph shown, each region proportion fluctuate at a certain level. For instance the percentage of first region of sample 1 fluctuate at around 83% which is a close value to the original proportion 81.44%. However, error bars or standard errors of each region become larger, as more SNPs are removed, which indicate that as more SNPs being removed, the proportions presented in ethnicity reports become less accurate. But in another case, the highest standard error for first and second region proportions of sample 1 and 2 are 1.32%, 1.41%, 1.33% and 1.03%. None of these standard errors exceed 1.5% which can be seen as an acceptable errors. Therefore the project concludes that when a large amount of SNPs are removed from a set of DNA data, the ethnicity report generated from the DNA data would be influenced, but the results are still acceptable to identify the owner's ethnicity.
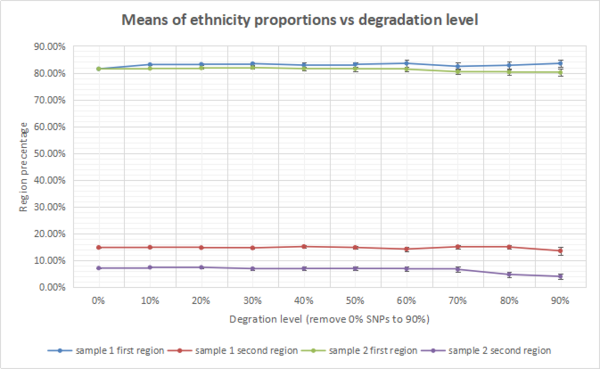
Conclusion[edit]
According to the observation of ethnicity change during the degradation process, as more amount of SNPs are removed from a complete human DNA reference file, the result of ethnicity report would be less accurate but the largest and second largest ethnicity regions in the report are still reliable. Therefore the top two major ethnicity of the Somerton Man are North Atlantic and Baltic, where these two regions are mostly around Europe.
Genetic diseases search[edit]
Aims[edit]
During this task, the team focused on searching clinical effects of each available SNP and identify any possible genetic disease or physical characteristics that Somerton Man could have.
Methods[edit]
To search the clinical effects of SNPs, the team developed a data mining program that collects information in SNP database. Python language was used for development since it is convenient for web development. The SNP database the project selected to use was dbSNP which is the largest database for nucleotide variations in the world, and is managed by the National Center for Biotechnology Information (NCBI). Figure 6.1 shows the information provided by dbSNP. The project team collected the clinical significance related to each rsid in Somerton Man's file.
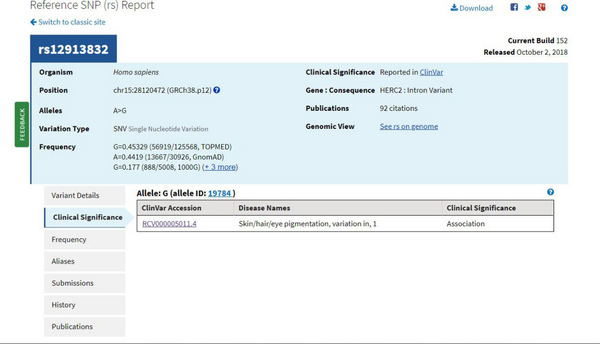
The program extracted every non-empty SNP in Somerton Man's DNA reference file. With the API provided by dbSNP, connection to dbSNP was established and each rsid of the extracted SNP was sent. When the connection was successfully set up, dbSNP sent back the information of corresponding SNP in JSON format. The data sent back was analysed and clinical information such as genetic disease name associated with the SNPs was recorded.
Result and discussion[edit]
With the support of data mining program, 613905 SNPs were searched in the database and 574 diseases were found. Figure 19 shows part of the genetic diseases outputs. As the figure shown, the program recorded the rsid of SNP that the disease belonged to in rsid column. dbSNP provides only a brief description of the clinical effects. More details are linked to another database called ClinVar which is a freely accessible, public database that provide medical reports of the relationships among human variants and phenotypes [12]. Therefore ClinVar Accession column is introduced to collect the ID of the recorded disease. This ID linked to the Clinvar database and allow the user to find a detailed medical report about the disease. The diseases names are recorded in disease name column. It is necessary to indicate that there are multiple diseases named with 'not specified' or 'not provided' which requires the user to find a detailed description of the disease in Clinvar. Unfortunately, none of diseases in the results relates or corresponds to Somerton Man's known characteristics.

Task 5: Investigation on DNA matches[edit]
Aims[edit]
The aims of this task is to investigate what results if the DNA match services provide on GEDmatch are conducted on high quality DNA kits, and how the degradation could effect the match results.
Methods[edit]
In task 2, the project has conducted DNA match examination on Somerton Man's DNA kit with multiple methods, but there is no match results for the his DNA reference file. In this task, one-to-many tool will be used again on 2 sample DNA files the project ordered from 23andMe, and the match results shall be recorded. Then the DNA match tests would be conducted on the degraded files created in task 3. The top 30 match results for each degraded DNA kit would be recorded and compare with the results of their original kit. A false positives and false negatives test would be conducted to show the change of match results during the degradation process. In this case, false positives would be match kits that are in the degraded kit's match results but not in original kit's result. And false negatives would present kits that are matched with original kit but not with the degraded one. An example is presented for a clear understanding. There are 5 kits A, B, C, D and E matched with the original kit, and kits A, B, C, M and N are matched with a degraded kit. Then the false positives for this degraded kit are D and E, and the false negatives are kits M and N. A line graphs of the number of false positives and false negatives against the percentage of SNPs removed will be created to show how degradation process effect the match results.
Results and discussion[edit]
Both DNA samples were successfully found their matched DNA kits in the database. Sample 1 have 8182 match kits and there are 5968 DNA files are found related to the sample 2. Top 30 match kits of sample 1 are shown in figure 20. The column Kit, Name and Email indicate the kit number, name of the kit and email of kit's owner. Column Total cM shows the total centimorgan which is a measure of genetic linkage between the 2 DNA kits. Note that the top 30 match kits are the kits with largest total centimorgan. Last but not least, the Overlap column present how many SNPs were used in the comparison between 2 kits.
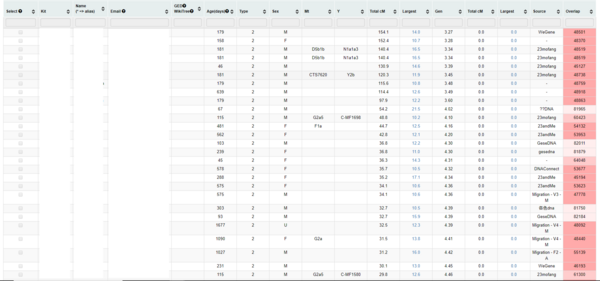
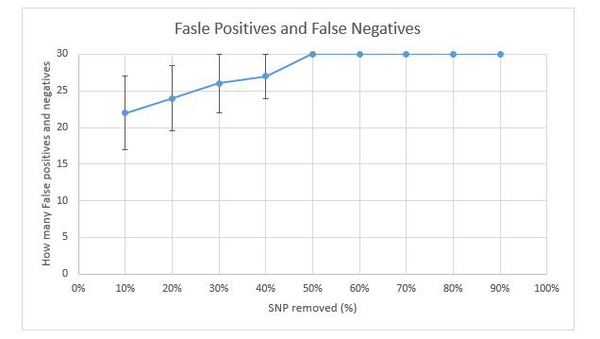
Next, the top 30 match kits for each degraded DNA reference files are recorded and the false negatives and false positives are calculated. Since all degraded files except the degraded files with 10% SNPs remaining have more than 30 match kits, the number of false negatives and false positives are same. The degraded files with 10% SNPs remaining have no match results. There are 4 degradation strategies introduced in task 3, therefore 4 sets of false negatives and false positives are provided for analysis. Figure 21 present line graph of the number of false negatives and false positives against degradation levels. The number of false negatives and false positives are the mean of 4 sets of data. Degradation level of 10% SNP remaining is not involved in the graph due to 0 match result. Similar graph which was done by last year's project were shown in figure 22. The DNA sample used in figure 22 is a completely different one from the samples used in figure 21. According to both graphs, the number of false positives and false negatives for different DNA samples are not same. But the trend are similar. As more SNPs are removed, the amount of false positives and false negatives increases until 50% SNPs are removed. When there is more than half amount of SNPs being removed, the number of false positives and false negatives reaches maximum of 30 which indicate that the match results of original kits and degraded kits are totally different at these levels. These results show that as more SNPs removed from the original DNA reference file, the match results would be more inaccuracy. And when there is only half amount of SNPs remaining in the DNA kit, the match results would be totally different and be unreliable. Moreover, when 10% of SNPs are removed, more than half of match results would be different which indicates that even a small amount of SNPs being removed could result a huge difference in DNA match test.
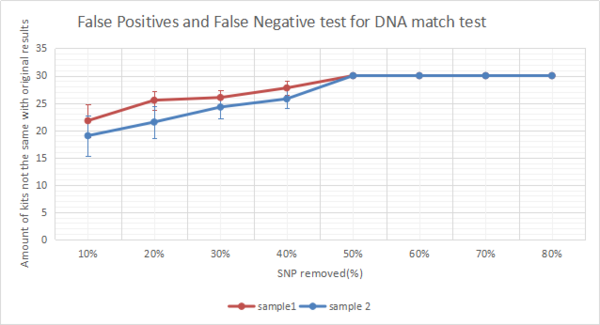
Conclusion[edit]
According to the findings in this task, the project can conclude that it would require a high level quality of DNA which would be at least more than 90% SNPs are available in the DNA reference file to receive a reliable DNA match results. Only a small amount of SNPs in the DNA file are changed could result a significant affect on DNA match results. In another case, if the Somerton Man's DNA reference file is available to be recovered to more than 20% SNPs remaining, there could be DNA kits found related to him. And If the Somerton Man's DNA kit could be recoverd to a level of 60% SNPs remaining, part of his DNA match results can be reliable.
Project Management[edit]
Budget[edit]
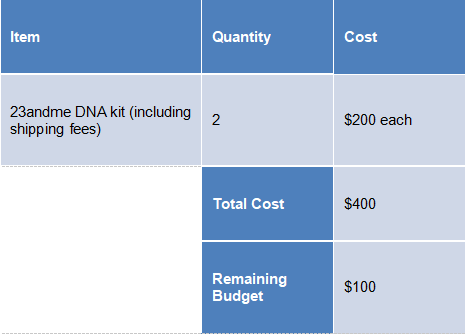
There are $250 budgets assigned to each member in the project, in which is $500 budgets in total for the project. Most budgets are spent on ordering 2 DNA kits from 23andme company for DNA testing. The details are shown in the table below. There is a plan on spending the rest of budgets on purchasing the advance services provided on GEDmatch. But the team is still evaluating demand of using these services.
Risk Management[edit]
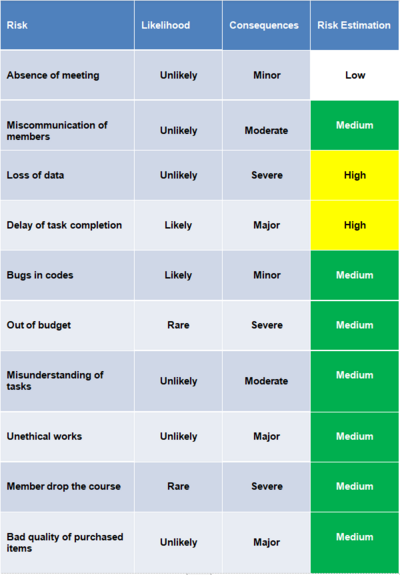
The risk assessment table are listed below. Several risks occurred during the progress. One of the group member was absent in the meeting several times due to time clash. But there is always at least one member attend the meeting with the supervisor. Members sometimes misunderstand assigned task, but issues were always fixed in the meeting in the following week.
Conclusions[edit]
The Somerton Man's DNA reference file provided to the project contain 613905 SNPs, but only 2.08% of SNPs that were not empty and were available for DNA analysis. With such low portion of available SNP, limited DNA analysis techniques can be conducted on the file. Unfortunately, there was no DNA kit that matched with Somerton Man's DNA kit found in GEDmatch database. According to the result of task 2 and task 4, the degradation process would have huge effect on the match results of a human DNA data. And it is impossible to recover Somerton Man's DNA by implementing simple recovery methods such as replacing empty SNPs with random base pairs or homozygous pairs. But if some reliable recovery strategies were introduced which have not been determined yet and allow Somerton Man's DNA to be recovered to more than 60% SNPs, then his relatives may be discovered. Moreover, the result from task 4 shows that the Somerton Man originated from Europe. To be specific, his ethnicity is about 36.21% North Atlantic and 20.44% Baltic. As for the genetic disease, 574 diseases were found, but there was no disease found that relates to his known appearance. So far, that is what the project can find in regards to the Somerton Man's DNA data. There was no clear clue that can lead to his identity. Who the Somerton Man is will still be a mystery.
Future Work[edit]
So far most work that can operated with the Somerton Man's DNA. Several points can be introduced to improve the outcome of the project. Firstly, there are only 2 DNA samples in this project for analysis. If more sample DNA reference files can be collected, then the analysis on degradation can be more reliable. Also, the ethnicity of 2 DNA samples are Asian, but the Somerton Man's ethnicity has higher chance to be European. Therefore, if the project can have several European DNA data, more reliable DNA analysis can be done. Moreover, another future job can be done by this project is to recover the Somerton Man's DNA data by inserting SNPs that are common among European. This could require a large amount of DNA samples for discovering common SNPs which could be a challenge. Last but not least, finding a higher quality DNA data of Somerton Man would be the best way to identify the man, since higher quality DNA can allow more genetic examinations to be conducted.
Reference[edit]
[1] Bineth, J, "Somerton Man: One of Australia's most baffling cold cases could be a step closer to being solved" This Is About, 13 December 2017. [online] Available at: https://www.abc.net.au/news/2017-12-14/somerton-man-cold-case-could-be- one-step-closer-to-solved/9245512 [Accessed 1 Jun. 2019].
[2] U.S. National Library of Medicine, "What is DNA?",U.S. National Library of Medicine, May. 28, 2019. [online] Available at: https://ghr.nlm.nih.gov/primer/basics/dna [Accessed 2 Jun. 2019].
[3] U.S. National Library of Medicine, "What is a chromosome?",U.S. National Library of Medicine, May. 28, 2019. [online] Available at: https://ghr.nlm.nih.gov/primer/basics/chromosome [Accessed 2 Jun. 2019].
[4] U.S. National Library of Medicine, "How many chromosomes do people have?",U.S. National Library of Medicine, May. 28, 2019. [Online]. Available: https://ghr.nlm.nih.gov/primer/basics/howmanychromosomes. [Accessed: 02- Jun- 2019].
[5] U.S. National Library of Medicine, "What are single nucleotide polymorphisms (SNPs)?",U.S. National Library of Medicine, May. 28, 2019. [Online]. Available: https://ghr.nlm.nih.gov/primer/genomicresearch/snp. [Accessed: 02- Jun- 2019].
[6] G. Shaw. “Polymorphism and Single nucleotide polymorphisms (SNPs)” Science Made Simple, Vol. 112, pp.664-665 2013.
[7] “DEAD MAN FOUND LYING ON SOMERTON BEACH” The News, December 1, 1948, p. 1 [online]. Available: https://trove.nla.gov.au/newspaper/article/129897161. [Accessed: 03- Jun- 2019].
[8] “Cryptic Note On Body” The News, June 6, 1949, p. 1 [online]. Available: https://trove.nla.gov.au/newspaper/article/36371152. [Accessed: 03- Jun- 2019].
[9] "Raw Data Technical Details", 23andMe, 2019. [Online]. Available: https://customercare.23andme.com/hc/en-us/articles/115004459928-Raw-Data- Technical-Details. [Accessed: 04- Jun- 2019].
[10] S. Zhang, "The Coming Wave of Murders Solved by Genealogy", The Atlantic, 2019. [Online]. Available: https://www.theatlantic.com/science/archive/2018/05/the-coming-wave-of- murders-solved-by-genealogy/560750/. [Accessed: 04- Jun- 2019].
[11] "General Information about dbSNP as a Database Resource", Center for Biotechnology Information (US), 2005. [Online]. Available: https://www.ncbi.nlm.nih.gov/books/NBK44469/. [Accessed: 06- Jun- 2019].
[12] Landrum, M., Lee, J., Riley, G., Jang, W., Rubinstein, W., Church, D. and Maglott, D. (2013). ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Research, 42(D1), pp.D980-D985.
[13] Chick, H. (2017). Finally! A Gedmatch Admixture Guide!. [Blog] genealogical musings. Available at: https://genealogical-musings.blogspot.com/2017/04/finally-gedmatch-admixture-guide.html [Accessed 29- Oct- 2019].
[14] Chen, J. and Seroka, A. (2018). Cipher cracking Final Report/Thesis 2018. [online] Eleceng.adelaide.edu.au. Available at: https://www.eleceng.adelaide.edu.au/personal/dabbott/wiki/index.php/Final_Report/Thesis_2018 [Accessed 1 Nov. 2019].
[15] Davidski. (2019). K13_population_averages. [online] Available at: https://docs.google.com/spreadsheets/d/1Oz6P5-SVEJciPX1TciGe-zoqA5JtOGIMG7nh-rCOj0c/edit#gid=804264822 [Accessed 1 Nov. 2019].
[16] Inside Story, presented by Stuart Littlemore, ABC TV, screened at 8 pm, Thursday, 24th August, 1978
Appendix A: Codes and tables used in the project[edit]
Appendix B: Tables used for the graph[edit]
- ethnicity line graph:File:Ethnicity changes.zip
- false positives and false negatives test: File:False.zip