Covid or flu? That is the question!
Supervisors
Research Project Students
Weekly Progress
The progress made each week by the members of the team, is recorded on this external web page. Moreover, any arising questions or concerns regarding the project supervisors are noted on this page. The Weekly Progress page will have its contents updated each week, until the completion of the project.
Project aim and scope
The Coronavirus Disease 2019 (COVID-19) is a respiratory infectious disease caused by the SARS-CoV-2 virus. The COVID-19 outbreak has a significant global impact on health care, economy and lifestyle. Non-COVID-19 viral pneumonia is also a common respiratory infection caused by viruses, bacteria or fungi. Chest radiography is capable of identifying patients with COVID-19 or non-COVID-19 viral pneumonia. Both diseases affect and damage the human lungs and causes various similar symptoms, which makes it a difficult task for differential diagnosis using chest X-ray images.
The aim of this project, ‘COVID or flu? That's the question!’, is to explore advanced techniques for image classification to determine whether chest X-ray images from patients that have COVID-19 versus those with non-COVID-19 viral pneumonia can be differentiated. This project will involve designing and constructing Machine Learning (ML) models, that can extract specific features from chest X-ray images and then learn from it, to perform accurate classification.
The scope of this project involves identifying and selecting numerous effective ML methods, that can be used to perform classification on chest X-ray images, within the specified given timeframe of this project. The ML model’s efficiency, relevancy to image classification problems, interpretability, and framework support availability, will all be analysed for selection.
The scope of this project involves identifying and selecting numerous effective ML methods, that can be used to perform classification on chest X-ray images, within the specified given timeframe of this project. The ML model’s efficiency, relevancy to image classification problems, interpretability, and framework support availability, will all be analysed for selection.
For this project, there will be two datasets that will be used. COVID-19 Radiography Database is a database created by researchers from various Asian universities. The journal articles, ‘Can AI Help in Screening Viral and COVID-19 Pneumonia?’ by M. E. H. Chowdhury et al and ‘Exploring the effect of image enhancement techniques on COVID-19 detection using chest Xray images’ by Rahman et al, has provided a ‘Data Availability Statement,’ allowing public use of their data [1] [2]. The dataset constitutes of 3616 COVID-19, 1,263 non-COVID-19 viral pneumonia and 10,192 healthy X-ray images.
The University of Montreal has provided a dataset that consists of 137 COVID-19, 90 nonCOVID-19 viral pneumonia and 90 healthy X-ray images. The journal article, ‘COVID-19 Image Data Collection: Prospective Predictions Are the Future’ by J. P. Cohen et al, also provided a 'Data Availability Statement,’ allowing public use of their data [3].
There exist many variants of the COVID-19 virus. However, for this project the scope is only limited to the SARS-CoV-2 strain of the coronavirus, which was first identified in Wuhan, China, in December 2019.
Background
According to World Health Organisation (WHO), there has been a total of approximately 452.2 million COVID-19 cases and 6.3 million deaths, as of March 14, 2022 [4]. The COVID-19 virus has caused an ongoing global pandemic and with newer variants being developed rapidly, the world is struggling to adapt. The first case of the COVID-19 virus was reported in December 2019, in Wuhan, Hubei Province, China, from where it began to transmit rapidly to the rest of the countries around the world [5].
For the diagnosis of COVID-19, various methods are used with the most common method being the Reverse transcription-polymerase chain reaction (RT-PCR) testing [6]. Although RT-PCR tests can be cost-effective, patients can expect a delay in testing and receiving results, especially during an outbreak. Numerous studies also concluded that RT-PCR testing has low sensitivity during the early stages of the infection, contributing to false-negative results [7] [8] [9].Chest imaging using X-rays and computer tomography (CT) scans are protocols currently practiced by healthcare centres to patients, that show strong respiratory symptoms [10]. Contrast to other popular methods such as RT-PCR testing and Rapid Antigen Testing (RAT), the process of using chest imagine is very simple, fast and provides greater accuracy due to its high sensitivity during the early stages of the infection [11]. Non-COVID-19 pneumonia is still one of the leading causes of death [12]. According to WHO, chest imaging using X-rays is the best method for diagnosing pneumonia [13]. Over the recent years, there has been a significant development in the areas of Artificial Intelligence (AI) and ML. With increasing computational power and growing amount of quality available data, ML methods especially using deep learning approaches, have shown good performances on medical imaging diagnosis [14].
The project contributes to the society mainly by providing an accurate differential diagnosis method to differentiate patients with COVID-19 and non-COVID pneumonia diseases. In comparison to non-COVID pneumonia, COVID-19 is highly transmissible and can display little to no symptoms, especially during the incubation period. Therefore, it is important to differentiate patients with COVID-19 and non-COVID pneumonia, to contain the spread of the COVID-19 virus and to assign appropriate medical treatments and measures. Both diseases display similar symptoms affecting the lungs and similar characteristics in the chest X-ray images. Figure 1 illustrates an example of a COVID-19, non-COVID-19 viral pneumonia and a healthy patient’s chest X-ray image.
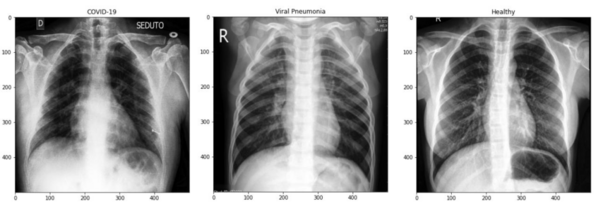
Hence, differential diagnosis using chest X-ray images can be tedious, even for expert radiologists. Therefore, this project provides an alternative method to health care centres, using ML models to improve their accuracy of diagnosing COVID-19 and non-COVID-19 pneumonia diseases using X-ray images. Additionally, burdens and stresses induced on health care staffs during an outbreak, when there is a significant influx of patients will be reduced, due to the provision of highly accurate automated processes.
A great detail of work has been published already involving the use of ML methods for medical imaging analysis. However, there are still areas of improvement in the analysis, as it requires proficiency and incorporates a diverse range of techniques to improve, accelerate and generate an accurate diagnosis. Several studies have showed that deep learning methods, more specifically, Convolutional Neural Networks (CNNs), have achieved better performance on image classification problems in comparison to other traditional ML models [15] [16] [17]. In this project, existing studies will be used as a guide to verify the work being conducted. Furthermore, this project will also focus on exploring novel ML techniques with the aim of improving the accuracy of differential diagnosis.
This project can prove to be highly valuable to the medical imaging field, as it will implement existing and novel, traditional and deep learning ML methods to accurately diagnose COVID-19 and non-COVID-19 pneumonia using chest X-ray images. The ML models will undergo a validation process, to fine tune its model parameters, which will allow it to generalise to unseen data. For this project, the validation methods, Hold-out Validation, k-Fold Cross-Validation (k-Fold CV) and Leave-one-out Cross-Validation (LOOCV) will be considered. The Hold-out Validation method involves splitting the training data into separate training and validation sets. For example, 60% of the training data could be used for training the model and the remaining 40% of the training data would be used for validating the model. The k-Fold CV method involves splitting the data into k folds, then training the data on k-1 folds and validating the data on the remaining fold. LOOCV is a variant of the k-Fold CV method, where each single sample is used for validation, while the remaining data is used for training the model. For both k-Fold CV and LOOCV, the average of all the individual evaluations is computed to determine the final result.
The ML methods will be tested using newly found datasets and evaluated using a series of evaluation metrics, to determine the best performing model. Performance measures such as Accuracy, Recall, F1-Score and Precision will be used. The work completed on this project can be extended to classify other common COVID-19 variants for accurate diagnosis.
Deliverables
- Project Plan
- Interim Seminar Slides
- Interim Progress Report
- Ingenuity Poster
- Ingenuity Video
- Final Report
References
- ↑ M. E. H. Chowdhury et al., "Can AI Help in Screening Viral and COVID-19 Pneumonia?," in IEEE Access, vol. 8, pp. 132665-132676, 2020, doi: 10.1109/ACCESS.2020.3010287.>
- ↑ T. Rahman et al., "Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images", Computers in Biology and Medicine, vol. 132, p. 104319, 2021. Available: 10.1016/j.compbiomed.2021.104319.>
- ↑ J. P. Cohen et al., "COVID-19 Images Data Collection: Prospective Predictions are the Future," J. Mach. Learn. Biomed. Imag., pp. 1-38, Dec. 2020.>
- ↑ "WHO Coronavirus (COVID-19) Dashboard", Covid19.who.int, 2022. [Online]. Available: https://covid19.who.int/ (Accessed: 14- Mar- 2022).>
- ↑ Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. (March 2020). "A pneumonia outbreak associated with a new coronavirus of probable bat origin". Nature. 579 (7798): 270–273.>
- ↑ Zoe C Brooks, ART, Saswati Das, MD, COVID-19 Testing: Impact of Prevalence, Sensitivity, and Specificity on Patient Risk and Cost, American Journal of Clinical Pathology, Volume 154, Issue 5, November 2020, Pages 575–584, https://doi.org/10.1093/ajcp/aqaa141.>
- ↑ Hellewell, J., Russell, T.W., Beale, R., Kelly, G., Houlihan, C., Nastouli, E. and Kucharski, A.J. (2021). Estimating the effectiveness of routine asymptomatic PCR testing at different frequencies for the detection of SARS-CoV-2 infections. BMC Medicine, 19(1).>
- ↑ S. Mallett et al., “At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? A systematic review of individual participant data,” BMC Medicine, vol. 18, no. 1, Nov. 2020, doi: 10.1186/s12916-020-01810-8.>
- ↑ Jarrom D, Elston L, Washington J, et al Effectiveness of tests to detect the presence of SARS-CoV-2 virus, and antibodies to SARS-CoV-2, to inform COVID-19 diagnosis: a rapid systematic review. BMJ Evidence-Based Medicine 2022;27:33-45.>
- ↑ Elsharkawy, M. et al. Early assessment of lung function in coronavirus patients using invariant markers from chest X-rays images. Sci. Rep. 11, 1–11 (2021).>
- ↑ X. Meng and Y. Liu, “Chest Imaging Tests versus RT-PCR Testing for COVID-19 Pneumonia: There Is No Best, Only a Better Fit,” Radiology, p. 203792, Oct. 2020, doi: 10.1148/radiol.2020203792.>
- ↑ Centers for Disease Control and Prevention, “Leading Causes of Death,” Centers for Disease Control and Prevention, 2019. https://www.cdc.gov/nchs/fastats/leading-causes-of-death.htm.>
- ↑ World Health Organization, Standardization of Interpretation of Chest Radiographs for the Diagnosis of Pneumonia in Children/World Health Organization Pneumonia Vaccine Trial Investigators’ Group, WHO, Geneva, Switzerland, 2001.>
- ↑ A. Anaya-Isaza, L. Mera-Jiménez, and M. Zequera-Diaz, “An overview of deep learning in medical imaging,” Informatics in Medicine Unlocked, vol. 26, p. 100723, 2021, doi: 10.1016/j.imu.2021.100723.>
- ↑ N. O' Mahony et al., “Deep Learning vs. Traditional Computer Vision.” [Online]. Available: https://arxiv.org/ftp/arxiv/papers/1910/1910.13796.pdf.>
- ↑ Yadav, S.S., Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J Big Data 6, 113 (2019). https://doi.org/10.1186/s40537-019-0276-2>
- ↑ Hedjazi, M.A., Kourbane, I. and Genc, Y., 2017, May. On identifying leaves: A comparison of CNN with classical ML methods. In 2017 25th Signal Processing and Communications Applications Conference (SIU) (pp. 1-4). IEEE.>